This article was published in Scientific American’s former blog network and reflects the views of the author, not necessarily those of Scientific American
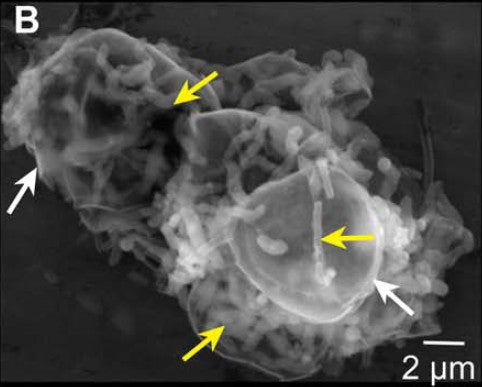
An agglomeration of Marinomonas bacteria(yellow arrows) and diatoms (white arrows). Credit: Guo et al. 2017
Sometimes your friends need a little persuasion to show up at social events you’ve worked hard to plan. The bacterium Marinomonas primoryensis doesn’t take any chances. It deploys long, sticky grappling hooks made of a single giant protein to rope its diatom and bacterial friends into socializing and then glue everyone to the underside of the ice floating atop their home. It knows that once you’re at the party, you’ll be totally glad you made the … er …effort.
That home is Antarctica’s
On supporting science journalism
If you're enjoying this article, consider supporting our award-winning journalism by subscribing. By purchasing a subscription you are helping to ensure the future of impactful stories about the discoveries and ideas shaping our world today.
Ace Lake, a salty, cold, stratified lake in the Vestfold Hills of Princess Elizabeth Land south of Australia. Ace Lake is very dark. Light penetrates only a few yards and oxygen no more than 40 feet. That’s a problem for some bacteria and all diatoms, microbes with intricate and beautiful glassy shells who require light to make food. It’s a particular problem for Marinomonas, which depends on the oxygen and nutrients produced by algae and diatoms to live.
Gravity being gravity, all these organisms will sink into the suffocating murk without some sort of assist. Antarctica being Antarctica, though, there is a ready-made life preserver floating right at the surface where the photons are maximal and the oxygen free-flowing, if only you could attach yourself to it somehow.
But gluing yourself, singular, to the ice would still present a problem: the surface is not a fixed target and constantly waxes and wanes. It would be all too easy to lose your grip on such a slippery life preserver. However, if you’re holding hands with scores of other bacteria, the odds that everyone will lose their grip at once are tiny.
The resulting “party” is actually a mixed-species community known as a
biofilm. Biofilms are ubiquitous in aqueous environments, from the pebbles on streambeds, to the tiles (and showerhead!) in your shower, to the film that you must scrub daily off your teeth. In the environments in which biofilms form, having a firm grip both on each other and on the surface you are filming upon is crucial to making it work. In Ace Lake, the result is a “symbiotic microcolony” where both bacteria and diatoms benefit from being neighbors in a sky-lit penthouse.
How these bacteria accomplish this feat is perhaps most amazing of all: they deploy massive ice anchors through portholes in their sides made of a single protein that measures 600 nanometers long (0.6 micrometers). That is as large as some
of the smallest bacteria and half the size of many others. The largest known protein – a human muscle protein named Titin – has a mass of 3.8 million Daltons, 244 protein folding domains, and a length exceeding 1 micrometer. The ice anchor, called somewhat awkwardly Marinomonas primoryensis ice-binding protein, has a mass of 1.5 million Daltons, and more than 130 protein folding domains. Unlike human muscle protein, which is found in organisms that are demonstrably much larger than Titin, the ice anchor is attached to an organism only about three times as long as the protein itself.
In a paper published in Science Advances in August, a team of Canadian, Dutch, and Israeli scientists report the full structure of the massive protein, which is divided into 5 major regions. The structures of Regions II and IV had been previously described, but I, III, and V were terra nova.

Overall structure of the Marinomonas ice binding protein. a) Domain map drawn to scale (aa = amino acids, the building blocks of protein). b) expanded view of RI and RIII to RIV. Percentages are the percent sequence similarity to the RII repeats c) Actual structures of various protein domains shown in cartoon form. OM = outer membrane, ligand = binding target. TISS = Type I secretion system, the targeting signal that sends the protein through the membrane pore which is a part of T1SS. RII has been greatly truncated in B and C as indicated by black crosshatches. Credit: Guo et al. 2017
After the ice anchor is synthesized, it somehow refrains from folding itself immediately. The entire giant protein finds a pore protein in the cell membrane and then threads the eye of the needle in its still-unfolded state. The scientists inferred this because the pore through which the protein most likely projects is too narrow to accommodate a folded protein. Once exposed to saltwater, calcium ions found there help fold the protein properly and become part of its structure.
What stops the protein from simply floating on out into the water? Region/Domain I contains a plug that is too big to fit through the pore and anchors the anchor inside the bacterium the way a button holds your shirt closed. Perhaps the plug is somehow folded prior to the rest of the protein.
Region II is a long chain of about 120 identical subunits that act as a boom, greatly extending the reach of these bacteria. It comprises 90% of the whole protein.
Region III comprises two docking ports that can bind both other Marinomonas bacteria and diatoms. One of the docks binds carbohydrates. It’s found in many other yeast and bacteria, and helps yeast perform a famous maneuver called flocculation that’s in one sense the opposite of what Marinomonas is trying to accomplish. When they run out of food, yeast bind to carbohydrates on each other’s exteriors using their dock. They coagulate and sink to the bottom of liquids because their collective weight increases. This survival strategy is only sporadically used by wild yeast, but
brewers have selected yeast strains for flocculation ability over the years to clear their beer before bottling.
The other dock binds to proteins and bears a resemblance to a dock found on Vibrio cholerae that helps it colonize the intestine of its human hosts.
Both docks likely bind other Marinomonas bacteria. Experiments by this team using Marinomonas docks engineered to glow in the dark show that they definitely bind to diatoms. When the scientists placed Marinomonas bacteria together with diatoms called Chaetoceros neogracile, the bacteria stuck to the diatoms (A second diatom species, Fragilariopsis cylindrus could not be captured by Marinomonas).
The whole mass ended up glued to any ice that was present in laboratory cultures too. Once lodged there, the wad of organisms resisted removal by flowing water. Diatoms by themselves showed no ability to stick to ice.
Region IV is the actual ice-anchor, which forms a panel built of amino acids that ingeniously arranges water molecules in an ice-like pattern. When this array of “pre-ice” touches actual ice, it freezes and anneals. Wow! That’s a big wow I think.
Region V, the first to encounter seawater as its being extruded from the bacterium, may help the rest of the protein fold properly. It also contains sequences that guide the unfolded protein to the proper pore.
This image created by the authors sums up their findings. The panel on the right is a bit hard to read so there's an enlargement below.
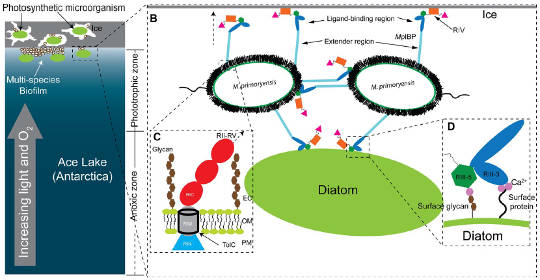
Credit: Guo et al. 2017
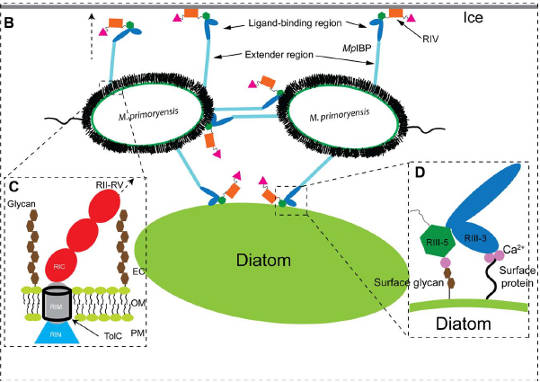
Model of M. primoryensis collectively binding with diatoms and other bacteria to ice. TolC is the membrane pore through which the ice anchor protein extends. Credit: Guo et al. 2017
The ice anchor as a whole bears a close chemical kinship to the proteins many pathogens use to form their own (from our perspective) more evil biofilms. These proteins, called Repeats-in-toxin adhesins, are used by notorious killers like Vibrio cholerae (cholera), Salmonella enterica (Salmonella), and some Pseudomonas bacteria to do their dirty work. They share the overall structure of the membrane anchor; the long, repetitive extender; and the binding and pore-targeting regions at its far end.
Despite the pathogens’ importance, not much is known about the details of their adhesins’ structures, how they stay attached to bacteria, and what biochemicals, exactly, they attach themselves to. The proteins’ enormous size and those pesky repeats make analysis difficult.
Knowing the structure of the Marinomonas ice anchor, the authors argue, may be the first step in figuring out how to deactivate these similarly-built grappling hooks that pathogenic bacteria use to colonize and exploit us. Indeed, scientists were able to produce antibodies that prevented Marinomonas from binding ice in the lab by jamming the ice anchor’s attachment plate. Crashing the party of some poor ice bacteria may seem lame, but it could eventually help us to shut down other fêtes so wild they are fatal to their hosts.
Reference
Guo, Shuaiqi, Corey A. Stevens, Tyler DR Vance, Luuk LC Olijve, Laurie A. Graham, Robert L. Campbell, Saeed R. Yazdi et al. "Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice." Science Advances 3, no. 8 (2017): e1701440.