This article was published in Scientific American’s former blog network and reflects the views of the author, not necessarily those of Scientific American
As we manage the COVID-19 catastrophe, we need to remember that in a world with an increasing global population, migrations, climate change and dramatic increases in global travel, such events are likely to occur more frequently. We need to consider the level of societal resources we’re prepared to spend to prevent future pandemics and identify the highest value strategies to invest in for their containment. Catastrophic outbreaks of infectious disease have occurred regularly throughout history. The transition of infectious diseases from animals to humans cannot be stopped.
But the spread can. The best way to prevent pandemics is to apply the same principles as we use to prevent catastrophic forest fires: survey aggressively for smaller brush fires and stomp them out immediately. We need to invest now in an integrated surveillance architecture designed to identify new outbreaks at the very early stages and rapidly invoke highly targeted containment responses.
Currently, we are not set up to do this very well. First, modern molecular diagnostic technologies detect only those infectious agents we already know exist; they come up blank when presented with a novel agent. Infections caused by new or unexpected pathogens are not identified until there are too many unexplained infections to ignore, which triggers hospitals to send samples to a public health lab that has more sophisticated capabilities. And even when previously unknown infectious agents are finally identified, it takes far too long to develop, validate and distribute tests for the new agent. By then, the window of time to contain an emerging infection will likely be past.
On supporting science journalism
If you're enjoying this article, consider supporting our award-winning journalism by subscribing. By purchasing a subscription you are helping to ensure the future of impactful stories about the discoveries and ideas shaping our world today.
But there is a strategy that could be implemented within a year at a reasonable cost. It would enable societies to halt the spread of emerging infectious diseases before they reach the pandemic stage. The concept combines real-time public health surveillance, next-generation sequencing technology and modern network communication and contact tracing.
Today, when sophisticated public health facilities around the world investigate clinical samples from an unexplained disease outbreak, they conduct a metagenomic analysis, comparing the patient samples to a database of genomic sequences of thousands of known infectious agents. It can identify both routine and unexpected infectious agents, even if the pathogen has never been seen before.
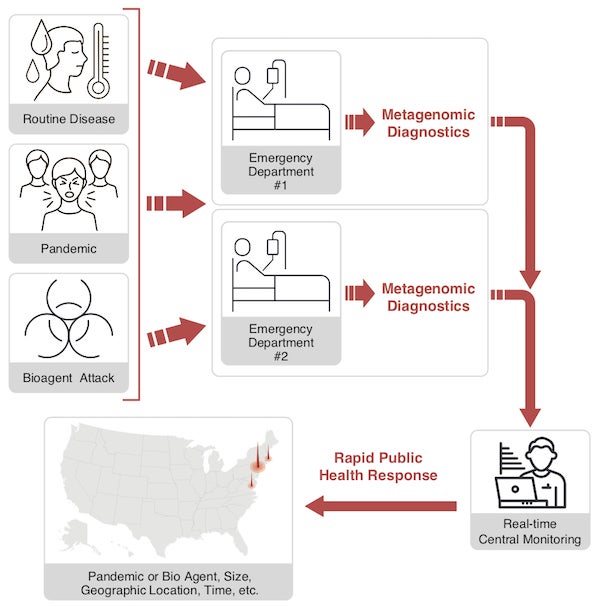
Proposed surveillance architecture for pandemic protection. Credit: Tracy Reigle
This type of analysis of COVID-19 specimens revealed that the causative agent was a virus similar to a previously known bat coronavirus. But in the new surveillance architecture proposed here, metagenomic analysis of patients presenting with severe clinical symptoms would be performed up front in hospital labs piggybacked on routine diagnostic testing. Although there are currently pros and cons regarding the use of metagenomic sequencing for primary diagnostics, the proposed architecture could be implemented at first as a surveillance system in parallel with conventional diagnostics and later as a primary diagnostic tool if warranted.
Large urban hospital emergency departments are the most strategically valuable locations to implement this surveillance architecture first, because these facilities capture a representative fraction of the patient population likely to be sickened by an infection relevant to public health. Given national epidemiological data—on infection rates; where symptomatic people seek health care; and how often diagnostic tests are ordered—a remarkably small number of surveillance sites would be required in order to identify an outbreak of an emerging agent.
A network of approximately 200 surveillance hospitals, located in U.S. metropolitan areas with populations of one million or more, would be needed to cover 30 percent of the emergency department visits in the nation. Monte Carlo models indicate a 95 percent probability of identifying an emerging infectious disease outbreak if only seven symptomatic patients seek health care in this system. Using metagenomic sequencing, the novel causative agent could be identified within hours, and the network would instantaneously connect the patients presenting at multiple locations as part of the same incident, providing situational awareness of the scope and scale of the event.
A practical example is the Washington, D.C., metropolitan area, home to about six million people. Based on national health care statistics, approximately 0.25 percent of the U.S. population seeks health care resulting in diagnosis with an infectious disease every day. Although only 20 percent of these patients will seek care in a hospital emergency department, it is reasonable to expect that most seriously ill patients who go first to physicians’ offices or urgent care centers will be ultimately sent to an emergency room. Health statistics show that in each large hospital emergency department, physicians order laboratory (respiratory or blood) tests for infectious disease on about 16 percent of the patients diagnosed with an infectious disease code.
Thus, in the Washington model, of the approximately 900 patients a day who present to an emergency department with a surveillance architecture, about 150 will be tested for infectious disease. When spread over just five D.C. hospital labs, this is an easily manageable number of daily metagenomic tests.
Following discovery of a novel outbreak, the key to managing it is the ability to answer four questions:
How many people are infected?
Where are they located?
When were the infections contracted?
With whom have these patients been in contact?
The computational infrastructure for answering these questions would be straightforward to develop because it is based on existing technology and statistical methods. The methods allow us to estimate of the size of an outbreak based on the number of presenting patients. The time of the initial contact with the infectious agent can also be estimated by working backward from reasonable assumptions about the incubation period. If this information is coupled with precise geolocation data volunteered by the patients from their cell phones, we can create a map of where patients are, updated in real time as each new case is added. Overlap of patient locations at the estimated time of infection could help identify the location and source. These data can also identify others who at risk for infection.
So how much time might be saved with the proposed surveillance architecture compared to the current system? The current methods we use to identify patients sickened with an agent of public health or security concern is based upon the “astute physician” model. That is, we depend on doctors’ clinical judgment to identify suspicious infections and send samples to a public health laboratory. The problem is that it is easy to miss an unusual event because symptoms from many different infections are very similar (think of the term flulike).
I asked a number of my colleagues in emergency medicine and public health how long they thought it would take to identify a novel, potentially dangerous infection. The response was highly variable, ranging from a few days to several weeks. There was consistent agreement that the answer is highly dependent on the infectious agent; the skill and experience of the physician; and the proximity and expertise of public health laboratory.
In a published study based on actual case presentations from the 2001 anthrax attack, only six of 164 physicians from civilian and military hospitals who reviewed the case reports suspected anthrax as a possibility. Thus, the performance of the current system is poorly understood, even by experts. However, most believe that, unless the infection occurred in a significantly large number of people and resulted in significant mortality, the event might take a very long time to identify, compared to a single day with the proposed surveillance architecture. Of course, in a geometrically progressing infection scenario, the early days are the ones that matter most for containment.
The strategy proposed here requires no new science and remarkably little cost to implement and maintain. The cost to set up and run a surveillance architecture in 200 urban hospitals in the US would be well under $1 billion, and it could be done within a year. This cost is beyond trivial compared to the current cost of the COVID-19 catastrophe. The CARES Act alone has cost our country over $2 trillion. It is also trivial compared to the amounts spent on preparedness for other catastrophic events like a conventional or nuclear war. Soon there will be many calls to invest more in public health infrastructure. I argue that we should first identify what a public-health infrastructure should look like in the 21st century and build a great one at the proper scale using the best available technology.
Read more about the coronavirus outbreak from Scientific American here. And read coverage from our international network of magazines here.